Creating better drugs to treat diseases is a challenging task. An ideal drug should attack the source of the illness and have no secondary effects. To this end, scientists first identify targets (e.g., a proteins) related to a disease, and the goal is to develop a drug that will interact with these targets (to cure the illness) but not with others (to minimize secondary effects).
This process traditionally involves screening millions of drug compounds (e.g. molecules) in search of a few ones that have the desired interactions with the targets. This produces a drug-target matrix of interactions. The screening process is expensive, so this matrix is highly incomplete.
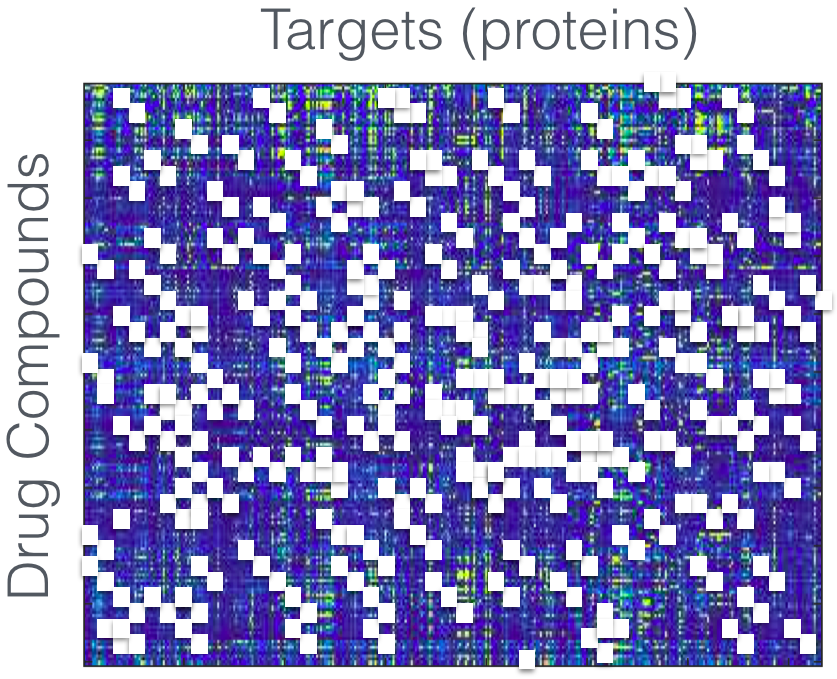
However, some targets behave similarly, so it is reasonable to assume that the interactions have some structure. For example a union of subspaces or a mixture matrix. Such structures may be exploited to predict the interactions of a new target. This way, given a new target, we may find its interaction with just a few cleverly selected drugs, see which targets have similar interactions, and predict the remaining interactions based on information about these similar targets.
© Daniel Pimentel-Alarcón